News
 Opal 3.0 Released
Opal 3.0 Released
We are pleased to announce that Opal 3.0 is now available. Opal is OBiBa’s core data management application for biobanks.
This release introduces a new major feature called Resources which is a novel way of interacting with existing data repositories: instead of importing data in a database that is managed by Opal, it is now possible to simply declare how to access the data (or a computation unit) with a formal representation called a resource. The access to the resource's data then happens directly in the R server managed by Opal. This feature uses the resourcer R package also developped by OBiBa and fully integrated with the DataSHIELD federated analysis platform. The resources open the possibility to make federated analysis of big data, complex and domain specific data structures (omics, geospatial etc.), using existing high performance computing facilities.
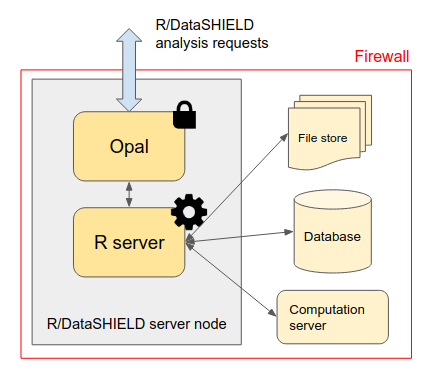
Additional features implemented:
- new Project python commands for creating/deleting a project and backup/restore its content have been added,
- user and group name auto-completion when adding permissions,
- integration wih Bioconductor R packages repository.
No breaking changes are expected with this release. It is even simpler to set up a new Opal system as no database is required when using only resources.
This release was possible thanks to the funding of the EUCAN-Connect program.
See Opal documentation for installation and operation instructions. We recommend to have a look at the section about Docker Image Installation .